Written By: Xiu Li
MB0902
092908H

A
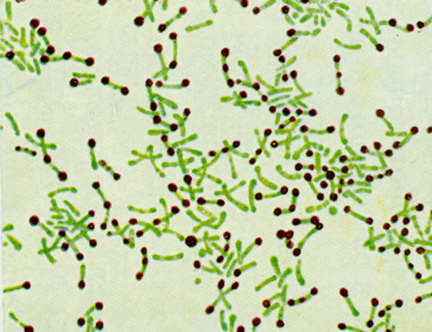
Flagellum is a tail-like structure that projects from the cell of certain cells. In this post, I'll be explaining Bacterial Flagella, as shown by the picture above.Flagella are made up of the protein flagellin and its shape is a 20 nanometer-thick hollow tube. They are helical filaments that have a sharp bend just outside the outer membrane, functions in locomotion and rotate like screws. The sharp bend, or "hook", allows the helix to point directly away from the cell.
A shaft runs between the hook and the basal body, passing through protein rings in the cell's membrane that act as bearings. Gram-positive organisms have 2 of these basal body rings, one in the peptidoglycan layer and one in the plasma membrane. Gram-negative organisms have 4 such rings: the L ring associates with the lipopolysaccharides, the P ring associates with peptidoglycan layer, the M ring is embedded in the plasma membrane, and the S ring is directly attached to the plasma membrane. The filament ends with a capping protein.

Video! : http://www.youtube.com/watch?v=0N09BIEzDlI
The bacterial flagellum is driven by a rotary engine made up of protein located at the flagellum's anchor point on the inner cell membrane. The engine is powered by proton motive force, i.e., by the flow of protons (hydrogen ions) across the bacterial cell membrane due to a concentration gradient set up by the cell's metabolism. The rotor transports protons across the membrane, and is turned in the process.
Flagella do not rotate at a constant speed but instead can increase or decrease their rotational speed in relation to the strength of the proton motive force. Flagellar rotation can move bacteria through liquid media at speeds of up to 60 cell lengths/second (sec). Although this is only about 0.00017 km/h (0.00011 mph), when comparing this speed with that of higher organisms in terms of number of lengths moved per second, it is extremely fast
The flagellar filament is the long helical screw that propels the bacterium when rotated by the motor, through the hook. In most bacteria that have been studied, the filament is made up of eleven protofilaments approximately parallel to the filament axis. Each protofilament is a series of tandem protein chains.
Through use of their flagella, bacteria who have them are able to move rapidly towards attractants and away from repellents. They do this by means of a Biased Random Walk (in which bacteria search for food or flee from harm), with 'runs' and 'tumbles' brought about by rotating the flagellum
Flagella arrangement schemesDifferent species of bacteria have different numbers and arrangements of flagella. There are basically four different types of flagellar arrangements.
1. A single flagellum can extend from one end of the cell - if so, the bacterium is said to be monotrichous.
2. A single flagellum (or multiple flagella) can extend from both ends of the cell - amphitrichous.
3. Several flagella (tuft) can extend from one end or both ends of the cell - lophotrichous; or,
4. Multiple flagella may be randomly distributed over the entire bacterial cell - peritrichous.
(amphi- a prefix meaning both or on both sides
lopho- or loph- a combining form meaning a "ridge" or "tuft,")

In some bacteria, the individual flagella are organized outside the cell body, helically twining about each other to form a thick structure called a fascicle. Other bacteria have a specialized type of flagellum called an "axial filament" that is located in the periplasmic space, the rotation of which causes the entire bacterium to move forward in a corkscrew-like motion.
Counterclockwise rotation of monotrichous polar flagella pushes the cell forward with the flagella trailing behind, much like a corkscrew moving inside cork. The flagella are left-handed helices, and bundle and rotate together only when rotating counterclockwise. When some of the rotors reverse direction, the flagella unwind and the cell starts "tumbling".
Such "tumbling" may happen occasionally, leading to the cell seemingly thrashing about in place, resulting in the reorientation of the cell. The clockwise rotation of a flagellum is suppressed by chemical compounds favorable to the cell (e.g. food), but the motor is highly adaptive to this. Therefore, when moving in a favorable direction, the concentration of the chemical attractant increases and "tumbles" are continually suppressed; however, when the cell's direction of motion is unfavorable (e.g., away from a chemical attractant), tumbles are no longer suppressed and occur much more often, with the chance that the cell will be thus reoriented in the correct direction.

(arrows are pointing to the Flagella)
References:
http://en.wikipedia.org/wiki/Flagellumhttp://www.life.umd.edu/classroom/bsci424/BSCI223WebSiteFiles/Flagella.htm